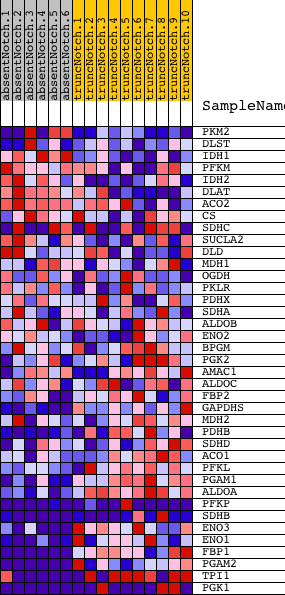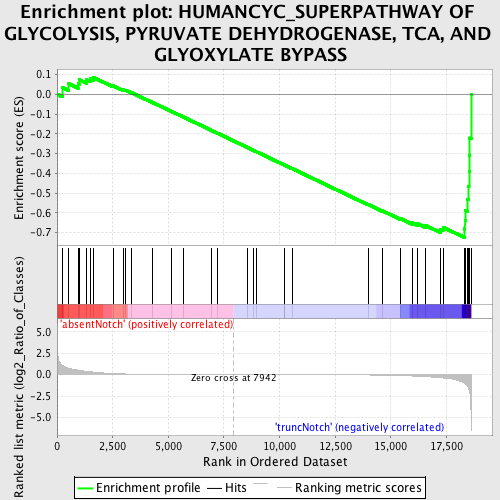
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | HUMANCYC_SUPERPATHWAY OF GLYCOLYSIS, PYRUVATE DEHYDROGENASE, TCA, AND GLYOXYLATE BYPASS |
| Enrichment Score (ES) | -0.7246548 |
| Normalized Enrichment Score (NES) | -1.5194074 |
| Nominal p-value | 0.017307693 |
| FDR q-value | 0.4210499 |
| FWER p-Value | 0.996 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PKM2 | 6520403 70500 | 229 | 1.062 | 0.0353 | No | ||
| 2 | DLST | 2320722 | 498 | 0.761 | 0.0551 | No | ||
| 3 | IDH1 | 1990021 | 943 | 0.524 | 0.0547 | No | ||
| 4 | PFKM | 1990156 5720168 | 991 | 0.504 | 0.0748 | No | ||
| 5 | IDH2 | 2680397 | 1321 | 0.381 | 0.0742 | No | ||
| 6 | DLAT | 430452 | 1488 | 0.338 | 0.0804 | No | ||
| 7 | ACO2 | 4230600 | 1644 | 0.293 | 0.0852 | No | ||
| 8 | CS | 5080600 | 2524 | 0.125 | 0.0435 | No | ||
| 9 | SDHC | 3140347 6100156 | 2970 | 0.082 | 0.0232 | No | ||
| 10 | SUCLA2 | 730632 1170292 | 3064 | 0.075 | 0.0216 | No | ||
| 11 | DLD | 4150403 6590341 | 3362 | 0.057 | 0.0082 | No | ||
| 12 | MDH1 | 6660358 6760731 | 4297 | 0.021 | -0.0411 | No | ||
| 13 | OGDH | 3840333 6350100 | 5120 | 0.011 | -0.0849 | No | ||
| 14 | PKLR | 1170400 2470114 | 5677 | 0.007 | -0.1145 | No | ||
| 15 | PDHX | 870315 | 6919 | 0.003 | -0.1812 | No | ||
| 16 | SDHA | 7000056 | 7190 | 0.002 | -0.1956 | No | ||
| 17 | ALDOB | 4730324 | 8552 | -0.001 | -0.2688 | No | ||
| 18 | ENO2 | 2320068 | 8815 | -0.002 | -0.2828 | No | ||
| 19 | BPGM | 5080520 | 8957 | -0.003 | -0.2903 | No | ||
| 20 | PGK2 | 2650091 | 10207 | -0.006 | -0.3573 | No | ||
| 21 | AMAC1 | 940270 | 10230 | -0.006 | -0.3582 | No | ||
| 22 | ALDOC | 450121 610427 | 10575 | -0.007 | -0.3763 | No | ||
| 23 | FBP2 | 1580193 | 13986 | -0.039 | -0.5582 | No | ||
| 24 | GAPDHS | 2690463 | 14608 | -0.056 | -0.5891 | No | ||
| 25 | MDH2 | 1850601 | 15423 | -0.097 | -0.6285 | No | ||
| 26 | PDHB | 70215 610086 | 15955 | -0.151 | -0.6503 | No | ||
| 27 | SDHD | 2570128 | 16214 | -0.186 | -0.6559 | No | ||
| 28 | ACO1 | 50114 | 16565 | -0.236 | -0.6642 | No | ||
| 29 | PFKL | 6200167 | 17218 | -0.351 | -0.6835 | Yes | ||
| 30 | PGAM1 | 2570133 | 17350 | -0.383 | -0.6734 | Yes | ||
| 31 | ALDOA | 6290672 | 18304 | -0.964 | -0.6814 | Yes | ||
| 32 | PFKP | 70138 6760040 1170278 | 18334 | -1.022 | -0.6371 | Yes | ||
| 33 | SDHB | 3120398 4780113 3710435 | 18367 | -1.094 | -0.5897 | Yes | ||
| 34 | ENO3 | 5270136 | 18458 | -1.372 | -0.5330 | Yes | ||
| 35 | ENO1 | 5340128 | 18494 | -1.566 | -0.4646 | Yes | ||
| 36 | FBP1 | 1470762 | 18514 | -1.688 | -0.3899 | Yes | ||
| 37 | PGAM2 | 3610605 | 18524 | -1.782 | -0.3104 | Yes | ||
| 38 | TPI1 | 1500215 2100154 | 18546 | -2.022 | -0.2208 | Yes | ||
| 39 | PGK1 | 1570494 630300 | 18610 | -5.004 | 0.0003 | Yes |